By the Visualization Core Lab team and Caitlin Clark
The KAUST Visualization Core Lab (KVL) enables insight into data—whether this is scientific data from simulations, large machine learning datasets, or sensor data from microscopes, gene sequencers and remotely deployed vehicles. Through collaboration, the KVL team augments the efforts and domain expertise of KAUST researchers by providing complimentary technical knowledge with exploratory visualization and analytic tools.
KVL’s multi-year collaboration with KAUST Distinguished Professor Pierre Magistretti and research scientist Corrado Calì's KAUST-École polytechnique fédérale de Lausanne (EPFL) Alliance for Integrative Modelling of Brain Energy Metabolism project—itself a collaboration with the Swiss Blue Brain Project (BBP)—exemplifies the benefits and opportunities of this approach.
Understanding the brain
In the BBP, scientists work on building detailed and biologically accurate digital reconstructions and simulations of the mouse brain using supercomputers. Through the project, they hope to create “a radically new approach for understanding the multi-level structure and function of the brain,” the BBP stated.
One of Calì and Magistretti’s contributions to the BBP was the production of 3D models of astrocytes, a type of glial cell found in the brain. Astrocytes help provide nutrients to neurons; support the blood-brain barrier; assist with neurotransmission; help repair nervous tissue after any injuries; and also play a role in neuronal development and plasticity.
Historically, it has been difficult to produce 3D astrocyte models for different reasons, including the cells’ complexity. Magistretti, Calì and colleagues used a special technique called high resolution serial block-face electron microscopy for biological tissues to image the cells, producing reconstructions of four astrocytes.

KAUST Professor Pierre Magistretti (right) and former KAUST research scientist Corrado Calì (left), now an assistant professor at the University of Torino, Italy, worked with the University’s Visualization Core Lab to understand the brain. File photos.
In a paper published on their work, they noted that “the imaged block was still too small to host the complete morphology of astrocytes and neurons, [but] the reconstructed volume still contained all the proximal processes of [the] cells.” The researchers then performed quantitative analyses on the models.
Using their 3D models, the Magistretti-Calì team worked to discover the mechanism through which lactate, an organic molecule produced by many of the body’s tissues, flows from astrocytes to neurons and not from neurons to astrocytes.
The team also examined how much of the lactate coming from glycogen, a simple sugar that provides energy to the brain, contributed to the astrocyte-neuron lactate shuttle hypothesis, which was previously developed by Magistretti and colleagues, and how the mechanism contributed to the use of energy by neurons. The research gives a big picture overview of how neurons and glial cells work together to coordinate brain energy metabolism, and the results have become part of the brain model in development with the BBP.
KVL expertise
For Magistretti and Calì's project, the KVL team provided systems expertise and access to the large memory and interactive computing resources needed to segment gigabyte-sized image stacks of brain tissue into cellular components.
The Magistretti-Calì team introduced KVL to ilastik, a unique image segmentation tool developed at the European Molecular Biology Laboratory. KVL then showed them how to use Avizo, a data visualization and analysis tool. It helped to assist them with skeletonization of the segmented cells into graphs and also enabled integration of the products with the rest of their tools and workflows.
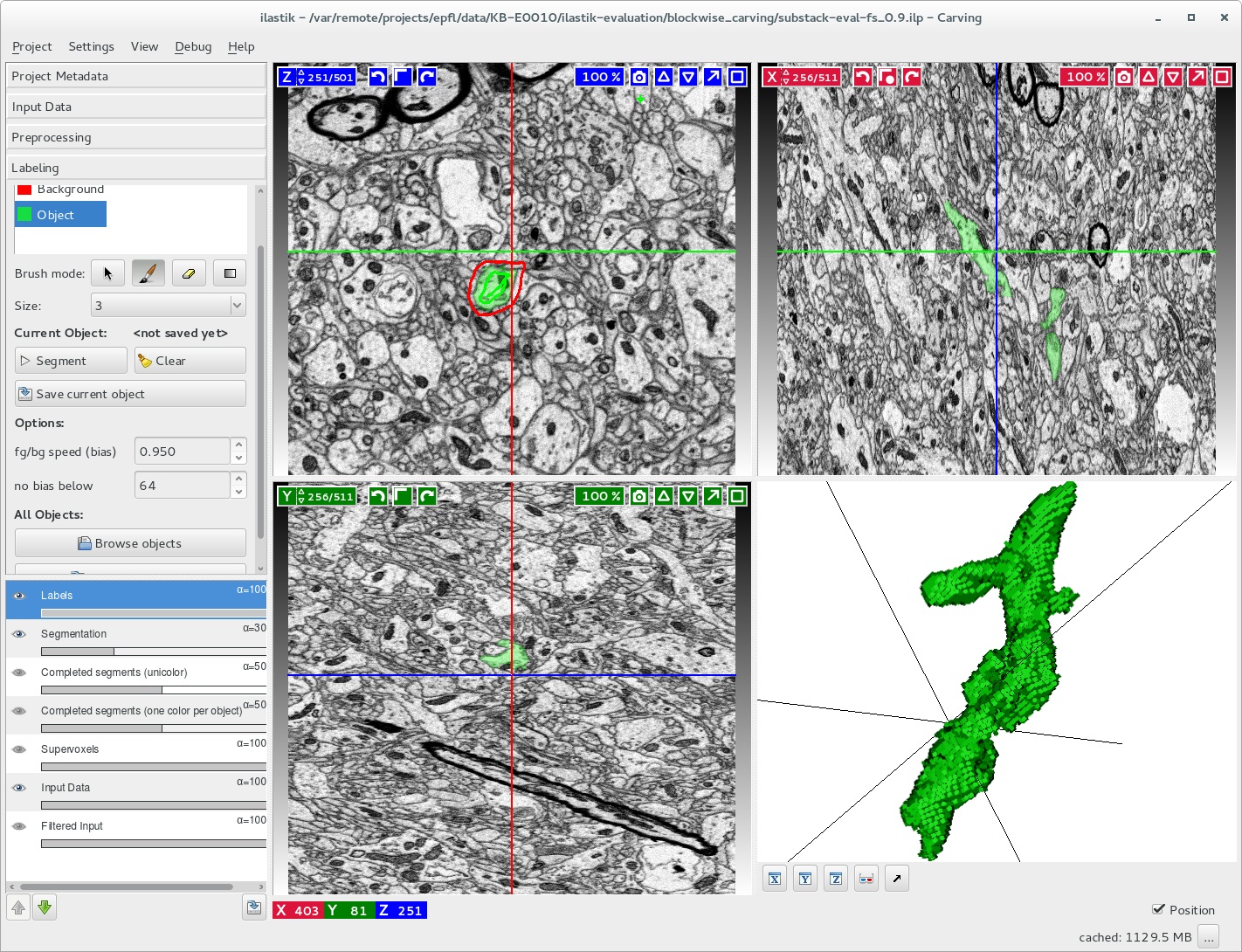
The KAUST Visualization Core Lab (KVL) team helped Prof. Pierre Magistretti and his team evaluate astrocyte segmentation in large image stacks using an enhanced version of ilastik, an image segmentation tool (pictured here). Image courtesy of KVL.
When image stacks from Magistretti and Calì became too big to process—even with the largest workstation—the KVL team applied its software development expertise to modify ilastik algorithms so that it continued to work. KVL’s experience with ilastik has given the team the ability to provide image segmentation capabilities on the Ibex compute cluster for all KAUST researchers.
“Upscaling the segmentation on large blocks was challenging, and the channel the KVL team opened with the ilastik team to work on the semiautomated segmentation pipeline was something we would not have been able to do fully independently,” noted Calì.
As the number of individuals working with Magistretti and Calì continued to grow, the biologists and segmentation technicians needed to review their work together. KVL’s large Showcase Display Wall enabled the entire team to see entire swaths of data clearly; work with familiar tools; and communicate directly.
“The Display Wall is a place where images and visualization help to bridge the language gaps of technical jargon,” said Glendon Holst, visualization scientist, KVL.

A segmented astrocyte is shown here in MeshLab, which is a processing system for 3D triangular meshes. Image courtesy of the KAUST Visualization Core Lab and Professor Pierre Magistretti.
Virtual reality collaborations
Virtual reality (VR) also played an important role in the Magistretti-Calì collaboration with KVL. VR presents 3D data in a way that preserves essential 3D spatial relationships, and it aided in the process of verifying alignment between the original image stack and that of the segmentation. Without VR, points that are far apart may appear nearby—as if they are in the same line of sight on a 2D monitor.
VR is also useful to explore dense, complicated structures, such as those found in the brain.
The Magistretti-Calì team collected a very high-resolution microscope image stack of a small region of rat hippocampus, part of the vertebrate brain, from EPFL in Switzerland. The image resolution from the microscopes was of such high quality that the biologists could make out individual granules of glycogen embedded in the segmented astrocytic cells. As the researchers sought to find out where the glycogen might be going, they faced challenges, such as the fact that the granules were frozen in time by the microscopy process.
“The problem of deciphering the complexity was like determining traffic flow in a city from just one satellite photograph—except, instead of a neat grid-like structure of roadways in 2D, there is a tangled mess of packed cells in 3D,” explained Holst. “Our solution was to put the biologists from the Magistretti-Calì team inside their data using a six-sided, fully immersive ‘Cave’ VR system.”
“There, the biologists could literally walk through the maze of cells together, seeing into the local neighborhood of cellular structures while collaboratively exploring their hunches,” he continued. “With the new insight, they were guided to the appropriate mathematical analysis to apply to their data.”

A researcher examines work on astrocytes in the unique KAUST ‘Cave’ Virtual Reality system. File photo.
Communicating science
The KVL team used Unity, a popular development platform for creating 2D and 3D interactive experiences—along with team members’ collective expertise in modeling, scientific visualization and software development—to create VirtualMente, a VR education experience that helps to communicate the work of the Magistretti-Calì team to a lay audience.
“With our assistance, the biologists now have many accurate 3D cellular models and a scientific story to tell,” Holst said.
“The development of VirtualMente was a project that would not have been possible without KVL, and it put together artistic skills to bring our science alive with VR programming,” added Magistretti.
“We continue to use Unity as the core of our VR capabilities,” noted Dina Garatly, visualization scientist, KVL. “Recent collaborations have used Unity to help understand flight data from near misses; highlight discoveries of complex enzymes; support exploration into agent-based reinforcement-learning frameworks; and provide simulation-based training for dangerous equipment.”

Visualization Core Lab scientists (from left to right) Dina Garatly, Sohaib Ghani and Glendon Holst assisted KAUST Professor Pierre Magistretti and colleagues with imaging the brain for their research. File photos.
Bringing researchers together
“Our work with KVL has been an exciting journey,” Magistretti said. “I think we mutually benefited from individual expertise over our years of collaboration. Our team received much technical input from the KVL team—especially on the programming side—and the KVL team was eager to learn about the brain. In discussing the challenges of our work, KVL was very interested in the science behind our analysis needs, rather than just listening to the bare problem. This helped to really build an infrastructure to develop the visualization and analysis tools and strategies that suited our needs.”
“We have several other datasets that we are still working on,” he continued. “One hot topic is to achieve a fully automated dense segmentation, meaning that we will be able to automatically label all processes and cells in our 3D stacks. Although we are independently working on this now, we could open a collaborative channel with KVL on the topic, as I believe the technique might be beneficial for many other scientific fields.”
“KVL helps bring people, facilities and workflows together through training and expertise,” stated Sohaib Ghani, lead visualization scientist, KVL. “The team offers training on scientific visualization and data science and analysis and best practices for deep learning. Feel free to start a conversation with us by emailing help@vis.kaust.edu.sa.”

